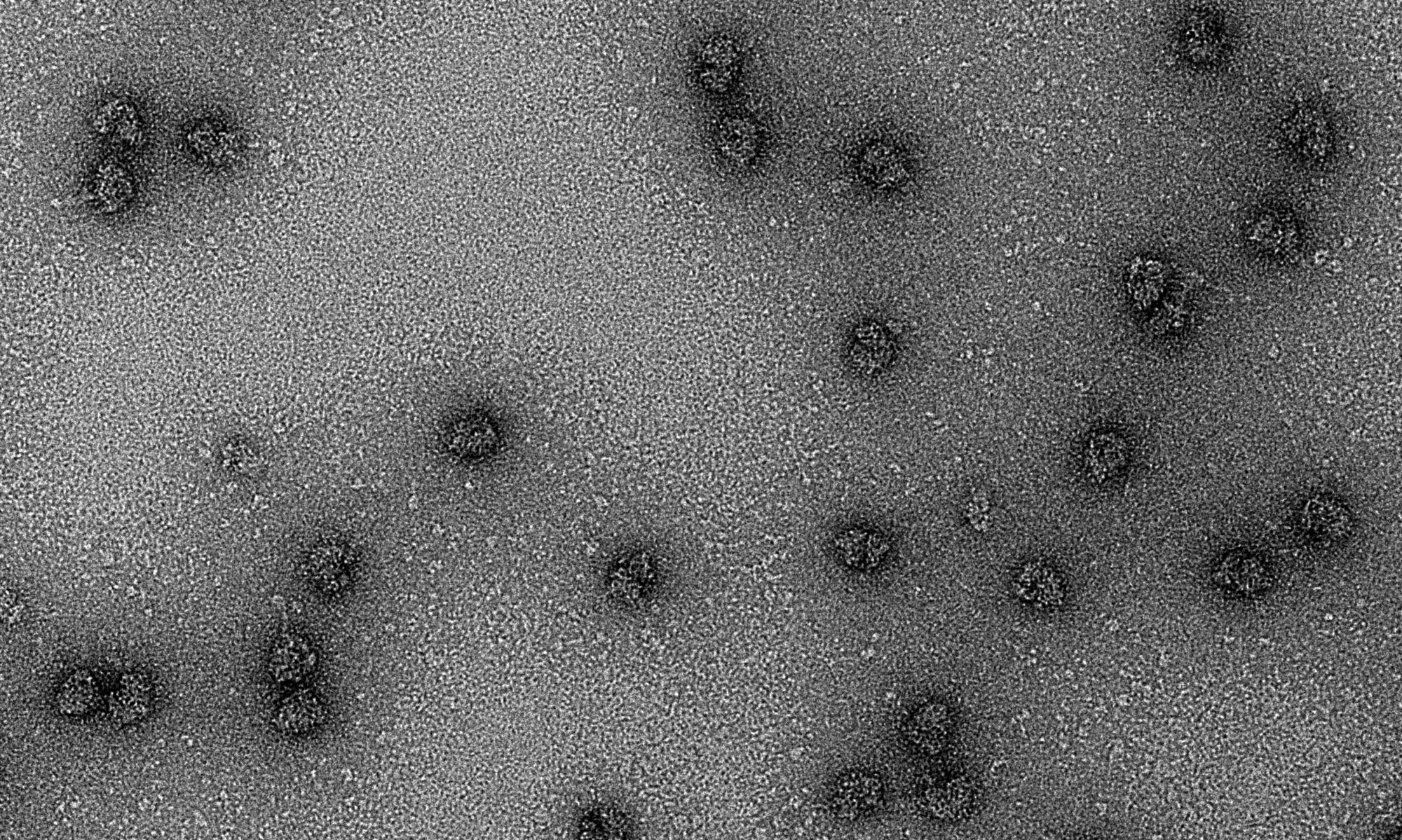
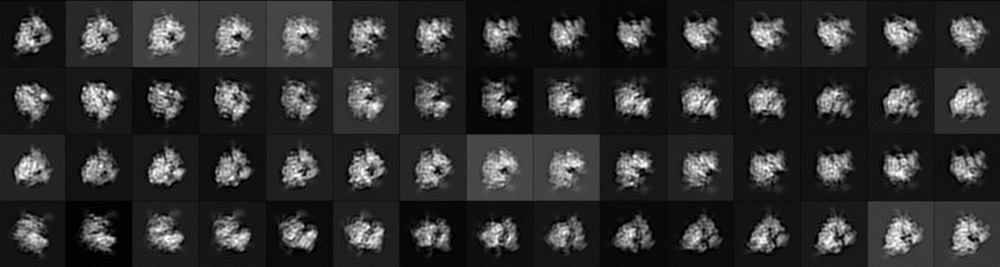
Research in the Filbin Lab focuses on the fundamental mechanisms and biomolecular structures that regulate eukaryotic translation initiation.
We are an inclusive lab where all are welcome! We believe in teamwork and the journey of discovery. Our all-undergraduate laboratory consists of members who diligently lead their own projects and work side-by-side with lab mates, creating a supportive research environment where everyone can be successful.
We celebrate the little successes along the winding research road, and we also celebrate each other – birthdays, awards, presentations, and graduations – because research is a human endeavor…and it’s the humans that matter the most!
Non-coding RNA Structure-Function
Non-coding RNAs (ncRNAs) are essential players in biology; they form the core of important cellular machines, they decode genetic information, they regulate gene expression and they functionally and structurally manipulate other macromolecules.
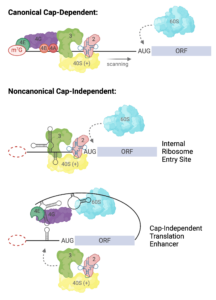 The family of ncRNAs we study are encoded in the 5′ and 3′ untranslated regions (UTRs) of viral genomic RNAs. These UTRs are capable of recruiting 80S ribosomes independent of the methylated guanosine cap structure found on the 5′ end of eukaryotic mRNAs, and often function in the absence of canonical translation initiation protein factors. Each virus has adapted to functionally replace the 5′ cap and host proteins factors via unique RNA structure-driven methods. By studying the structure-based mechanism of translation initiation, we aim to learn about fundamental mechanisms of eukaryotic translation initiation and the roles of lncRNAs in biology.
The family of ncRNAs we study are encoded in the 5′ and 3′ untranslated regions (UTRs) of viral genomic RNAs. These UTRs are capable of recruiting 80S ribosomes independent of the methylated guanosine cap structure found on the 5′ end of eukaryotic mRNAs, and often function in the absence of canonical translation initiation protein factors. Each virus has adapted to functionally replace the 5′ cap and host proteins factors via unique RNA structure-driven methods. By studying the structure-based mechanism of translation initiation, we aim to learn about fundamental mechanisms of eukaryotic translation initiation and the roles of lncRNAs in biology.
Translational Regulation in Developing Neurons
Protein synthesis is an indispensable yet energetically costly process within the cell. Considering this, translation is tightly coordinated with the condition of the cell. Both intra- and extracellular signals dictate when translation occurs as well as which proteins are produced based on current cellular needs. During cellular development, protein synthesis is spatiotemporally regulated to ensure proper tissue architecture formation.
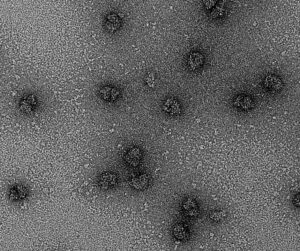 Neurons are highly polarized cells that develop directionally in response to environmental chemotropic factors. Part of this response involves tethering and releasing translation machinery at axonal growth cones pending the chemotropic signal received. By studying the structure and biochemical makeup of these translation complexes, we aim to shed light on how protein machinery can be manipulated during cell and tissue development.
Neurons are highly polarized cells that develop directionally in response to environmental chemotropic factors. Part of this response involves tethering and releasing translation machinery at axonal growth cones pending the chemotropic signal received. By studying the structure and biochemical makeup of these translation complexes, we aim to shed light on how protein machinery can be manipulated during cell and tissue development.
Designing Novel SHAPE Probes
 Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) is a biochemical modification assay that identifies RNA flexibility that often coincides with single-stranded nucleotides. The half-life, solubility, and reactivity of SHAPE probes vary providing us with a diverse biochemical toolkit to measure RNA structural dynamics. In collaboration with Dr. Shailesh Ambre at MSU Denver, we are synthesizing novel SHAPE probes that provide users with a more flexible platform to perform in vitro and in-cell RNA structure probing.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) is a biochemical modification assay that identifies RNA flexibility that often coincides with single-stranded nucleotides. The half-life, solubility, and reactivity of SHAPE probes vary providing us with a diverse biochemical toolkit to measure RNA structural dynamics. In collaboration with Dr. Shailesh Ambre at MSU Denver, we are synthesizing novel SHAPE probes that provide users with a more flexible platform to perform in vitro and in-cell RNA structure probing.
CURE & Near-Peer Mentoring Effects on Student STEM Identity
Course-based undergraduate research experiences (CUREs) provide students with genuine research experiences, exploring novel and relevant hypotheses through iteration and often collaboration. These courses are typically offered to freshmen/sophomore-level students with the goal of increasing retention-to-degree. We designed a novel CURE (though the NSF-funded CUREnet) focused on providing these experiences to senior-level students with a special emphasis on mentoring and career exploration to increase retention-to-career.

We are studying the effects of a new CURE on student self-perception as scientists, confidence and feelings of belonging in STEM, as well as the evolution of their career aspirations through the course. In partnership with the RNA Bioscience Initiative at the University of Colorado, School of Medicine, we are analyzing these effects in both undergraduates and near-peer graduate student/staff mentors.
Collaborations
Research in our lab is performed in collaboration with the labs of Dr. Shailesh Ambre at MSU Denver, Drs. Kirk C. Hansen, and Beat Vögeli at the University of Colorado School of Medicine, as well as Dr. Janet Filbin at the University of Colorado Denver, School of Education and Human Development.
We are always open to new collaborations to foster a rich experience for our undergraduates as well as continue exciting research. Please contact us if you are interested in a joint project!